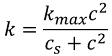Here are the M-files to implement composite trapezoidal rule for equally spaced data and unequally spaced data.
Hello, everyone! Welcome to Chemical Engineering World! In this blog, I will share my solutions of the practice and exercise of some of the Chemical Engineering subjects. I hope that it benefits students who struggle in the study of the subjects. Do check it out and stay tuned! 😊 - Learn smart, study hard
Tuesday, August 24, 2021
Numerical Integration Formulas
Numerical Integration Formulas
Problem 19.13.
Solution:
Numerical Integration Formulas
Problem 19.5.
Solution:
Saturday, August 21, 2021
Polynomial Interpolation: Inverse Interpolation
Problem 17.9.
Solution:
Polynomial Interpolation
Problem 17.5.
Solution:
Thursday, August 19, 2021
Polynomial Interpolation
Problem 17.4.
Solution:
Wednesday, August 18, 2021
Polynomial Interpolation
Here are the M-files to implement Newton interpolation and Lagrange interpolation.
Newton Interpolation M-file
% Newtint: Newton interpolating polynomial
% yint = Newtint(x,y,xx): Uses an (n - 1)-order Newton
% interpolating polynomial based on n data points (x, y)
% to determine a value of the dependent variable (yint)
% at a given value of the independent variable, xx.
% input:
% x = independent variable
% y = dependent variable
% xx = value of independent variable at which
% interpolation is calculated
% output:
% yint = interpolated value of dependent variable
% compute the finite divided differences in the form of a
% difference table
n = length(x);
if length(y)~=n, error('x and y must be same length'); end
b = zeros(n,n);
% assign dependent variables to the first column of b.
b(:,1) = y(:); % the (:) ensures that y is a column vector.
for j = 2:n
for i = 1:n-j+1
b(i,j) = (b(i+1,j-1)-b(i,j-1))/(x(i+j-1)-x(i));
end
end
% use the finite divided differences to interpolate
xt = 1;
yint = b(1,1);
for j = 1:n-1
xt = xt*(xx-x(j));
yint = yint+b(1,j+1)*xt;
end
Lagrange Interpolation M-file
Tuesday, August 17, 2021
Nonlinear Regression
Problem 15.11.
Solution:
General Linear Least-Squares Regression
Problem 15.10.
Solution:
Sunday, August 15, 2021
General Linear Least-Squares Regression
Here are the M-files to implement quadratic (second-order polynomial) regression, multiple linear regression and general linear least-squares regression.
Quadratic Regression M-file
Multiple Linear Regression M-file
General Linear Regression M-file
Wednesday, August 11, 2021
General Linear Least-Squares
Problem 15.6.
Sunday, August 8, 2021
Gauss Elimination
Here are the M-files to implement naive Gauss Elimination and Gauss Elimination with partial pivoting.
Naive Gauss Elimination M-file
Gauss Elimination with Partial Pivoting M-file
Friday, August 6, 2021
Linear Least-Squares Regression
Problem 14.14.
Solution:
Thursday, August 5, 2021
Linear Least-Squares Regression
Here is an M-file to implement linear regression.
Linear Regression M-file
Linear Least-Squares Regression
Problem 14.9.
Solution:
Numerical Integration Formulas
Here are the M-files to implement composite trapezoidal rule for equally spaced data and unequally spaced data. Composite Trapezoidal Rule ...
-
Problem 15.6. Use multiple linear regression to derive a predictive equation for dissolved oxygen concentration as a function of temperatur...
-
Here are the M-files to implement naive Gauss Elimination and Gauss Elimination with partial pivoting . Naive Gauss Elimination M-file func...
-
Problem 17.4. Given the data (a) Calculate f(3.4) using Newton's interpolating polynomials of order 1 through 3. Choose the sequence o...